Alex Johnson
Assistant Professor of Biochemistry 
Research Description
Mechanisms and evolution of cell death
A common strategy used by organisms in the defense against pathogens and cancer is the controlled self-destruction of compromised cells. These processes of programmed cell death are incredibly abundant and diverse in nature, being utilized by multicellular and unicellular organisms alike. To achieve success and spare healthy cells, programmed cell death must be rapid, tightly controlled, and tunable. Our laboratory is interested in how the molecular machines of cell death work. We seek to understand when these machines originated during evolution, how they sense pathogen infection, how they rapidly eliminate infected cells, and how they alert surrounding cells. To answer these questions, we employ a diverse range of techniques including biochemistry, microbiology, and structural biology.
Pore-forming proteins and the evolution of pyroptosis
Membrane pore-forming proteins are central effectors of the mammalian programmed cell death processes apoptosis, necroptosis, and pyroptosis. Previously, I discovered that gasdermin pore-forming proteins, the defining molecules of pyroptosis, are conserved in bacteria where they execute host cell death and defend against the viruses of bacteria—bacteriophages. By applying X-ray crystallography and single-particle cryogenic electron microscopy (cryo-EM), I defined the complete mechanism of bacterial gasdermin pore formation and explained how an evolutionarily conserved lipid modification mediates this process. My research revealed a deep conservation of the cell death machinery across life and established exciting new perspectives on mammalian pyroptosis.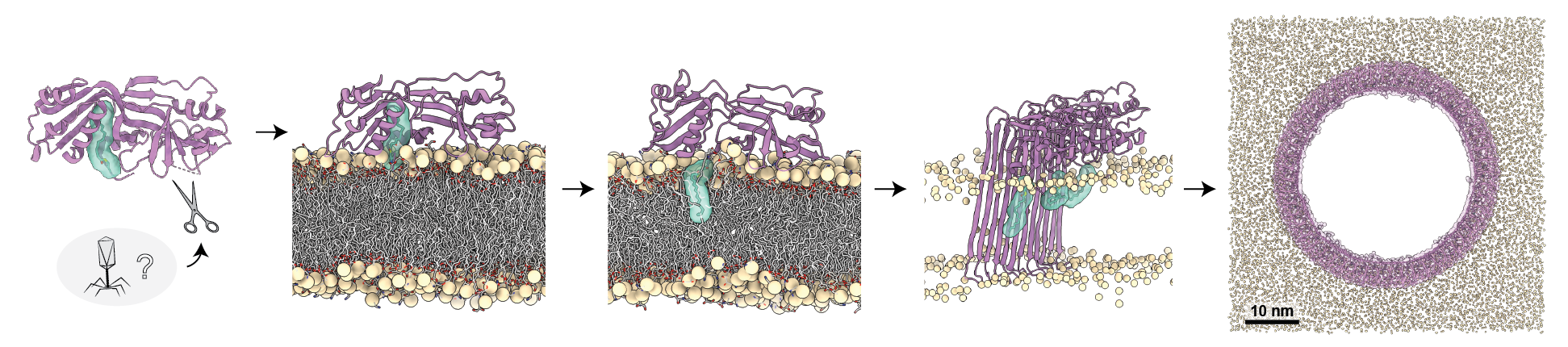
Building on these discoveries, our laboratory seeks to explain how cell death machinery controls host-pathogen interactions and define new mechanisms pore-forming proteins in host–pathogen conflict. We are specifically interested in how these shape-shifting proteins are triggered into activation, how they target lipid membranes of diverse composition, and how they oligomerize into gigantic membrane pores. To establish the rules by which these fascinating proteins work, we are focused on unique candidate proteins across life. A long-term goal is to apply our knowledge of pore-forming proteins to engineer novel protein-based therapeutics and functional materials.
Mechanisms of pathogen sensing and evasion across life
Immune systems must be exquisitely selective in discriminating self from non-self. This is particularly the case for programmed cell death, where incorrect sensing has fatal consequences for the cell and can lead to inflammation for the organism. In multiple cell death signaling pathways, proteases and protease complexes serve as the sensors that initiate cell death upon pathogen sensing. We are interested in how these molecules detect pathogen- or infection-derived molecules, how they recognize their downstream substrates, how they oligomerize into supramolecular complexes, and how they use energy in the form of ATP. Additionally, we are interested in how pathogens evade immune detection and the potential for such strategies in anti-inflammatory drug development. Our laboratory is using microbiology, biochemistry, and cryo-EM to address these problems.
Selected Publications
Johnson, AG; Mayer, ML; Schaefer, S; McNamara-Bordewick, N; Hummer, G; Kranzusch, PJ “Structure and assembly of a bacterial gasdermin pore,” Nature, 2024, 628: 657–663.
Boys, IN; Johnson, AG; Quinlan, M; Kranzusch, PJ; Elde, N “Structural homology screens reveal poxvirus-encoded proteins impacting inflammasome-mediated defenses,” Cell Reports, 2023, 42(8): 112878.
Johnson, AG & Kranzusch, PJ “What bacterial cell death teaches us about life,” PLOS Pathogens, 2022, 18(10): e1010879.
Johnson, AG; Wein, T; Mayer, ML; Duncan-Lowey, B; Yirmiya, E; Amitai, G; Sorek, R; Kranzusch, PJ “Bacterial gasdermins reveal an ancient mechanism of cell death,” Science, 2022, 375: 221–225.
Wang, J; Johnson, AG; Lapointe, CP; Choi, J; Prabhakar, A; Petrov, AN; Puglisi, JD “eIF5B gates the transition from translation initiation into elongation,” Nature, 2019, 573: 605–608.
Johnson, AG; Lapointe, CP; Wang, J; Corsepius, N; Choi, J; Fuchs, G; Puglisi, JD “RACK1 on and off the ribosome,” RNA, 2019, 25: 881–895.